QDD
for LINUX and WindowsThis page is not updated any more.
Version 2.1 (28 June 2011)
The new web page of QDD is at
net.imbe.fr/~emeglecz/qdd
Emese Meglécz1 and Jean-François Martin2
1Aix-Marseille Université, CNRS, IRD, UMR 7263
– IIMBE, Equipe EGE Case 36, 3 Place Victor Hugo, 13331 Marseille
Cedex 3, France
2Montpellier SupAgro, INRA, CIRAD, IRD, Centre de
Biologie et de Gestion des Populations, Campus International de
Baillarguet, CS30016, 34988 Montferrier-sur-Lez, France
emese.meglecz(at)imbe.fr
http://www.univ-provence.fr/gsite/Local/egee/dir/meglecz/QDD.html
The original QDD (QDD1 hereafter) has been updated.
QDD2_beta is available in command line version both for Linux and Windows.
The GUI interface for windows is in preparation.
We strongly suggest that you use QDD2_beta. If you really insist on
using QDD1, BEWARE, some of the target microsatellites might be
redundant between the perfect and the compound microsatellite tables.
Reference:Meglécz, E., Costedoat, C., Dubut, V., Gilles, A., Malausa, T., Pech, N., Martin, J.F. 2010. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 2010; Vol. 26,3 : 403-404; doi: 10.1093/bioinformatics/btp670. LINK |
|||
Overview:Large scale sequencing has become affordable; therefore it is likely to replace rapidly microsatellite isolation involving cloning. Apart from cost- and labour-efficiency, access to a large number of sequences has two great advantages:
QDD treats all bioinformatics steps from raw sequences until obtaining PCR primers: sorting sequences by tag, removing adapters/vectors, detection of microsatellites, detection of redundancy/possible mobile element association, selection of sequences with target microsatellites and primer design. A user-friendly windows interface i-QDD2 is under development. The current version can be run both under Linux and Windows in an easy to use command line option. QDD1 can be run both under Linux and Windows in an a easy to use command line option, the windows version is supported by a Graphical User Interface called iQDD, designed and programmed by Frédéric Calendini. |
|||
Changes between QDD1 and QDD2:
|
|||
Installation:QDD is written in Perl and can be run as a standalone application on Windows or Linux systems.For both versions the following freely available programs should be installed in order to be able to run QDD: ActivePerl (http://www.activestate.com/activeperl/) BLAST (ftp://ftp.ncbi.nih.gov/blast/executables/) Use BLAST programs for QDD1 and BLAST+ for QDD2 ClustalW (Higgins et al., 1991, Larkin et al. 2007; ftp://ftp.ebi.ac.uk/pub/software/clustalw2/) Primer3 (Rozen, 2000; http://primer3.sourceforge.net/) Use version 1 (Primer3-1.1.4 for example) QDD1 users only : For Windows users, the above three programs can be downloaded directly from : http://www1.montpellier.inra.fr/CBGP/NGS/Files/softwares/blast-2.2.18-ia32-win32.exe http://www1.montpellier.inra.fr/CBGP/NGS/Files/softwares/clustalw-2.0.12-win.msi http://www1.montpellier.inra.fr/CBGP/NGS/Files/softwares/primer3-1.1.4-WINXP.zip Our program has been tested thoroughly with these versions. QDD2 users : Install Bioperl ( http://www.bioperl.org/ ) It is only necessary for the last step of BLASTing sequences to Benank (pipe4) Help for windows installation http://www.bioperl.org/wiki/Installing_Bioperl_on_Windows ) For the command line installation of QDD see the documentation. The installation of the Windows version (iQDD) is straightforward but requires that the Microsoft® .NET 3.5 Framework is installed on your computer. This is usually the case when Windows updates are done regularly but it may be absent from older systems. The .NET framework 3.5 package is available for free here and should be installed prior to iQDD installation. When first launched, iQDD will ask you to enter the full path of the external applications used (Clustalw2, Blast and primer3) on your hard drive. |
|||
|
Higgins DG, Bleasby AJ, Fuchs R (1991) CLUSTAL V: improved software for multiple sequence alignment. Cabios, 8, 189-191. Larkin,M.A., Blackshields,G., Brown,N.P., Chenna,R., McGettigan,P.A., McWilliam H., Valentin,F., Wallace,I.M., Wilm,A., Lopez,R., Thompson,J.D., Gibson,T.J. and Higgins,D.G. (2007) ClustalW and ClustalX version 2. Bioinformatics 23(21) 2947-2948. Rozen,S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386.
|
|||
Quick start guide:The use of the command-line version is both available for LINUX and Windows operating systems and is fully detailed in the manual found here.The Graphical user interface is available for Windows and we detail here the first critical steps to get started with QDD with this solution in mind. After installation, which should be straigthforward, the user needs to define the paths for external applications such as Clustalw2, Blast and Primer3. Those external applications should already be installed on your system and are freely available (see installation section for URLs). When first running the iQDD Graphical Interface (Launch iQDD from the Start menu), the application will open the « Options » window (you can access this dialog later by going to the « Options » menu and select « Preferences »). This window will allow you to define the paths for external applications through a folder browser (see screenshot below). 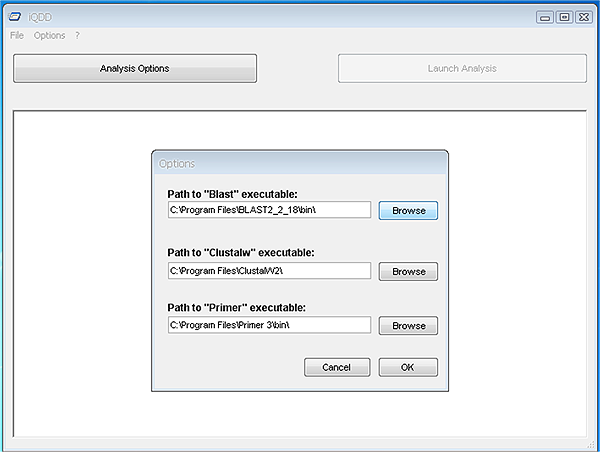 Note: The executables for Blast and Primer are found in their « bin » folders, while the executable for Clustalw2 is directly located at the root of its installation folder. The second step is to define the analysis options. It is done by selecting the corresponding button on the main window. The « Analysis settings » window opens and you can redefine the parameters that best fit your needs. The only obligatory field is the path to the data (see screenshot below). 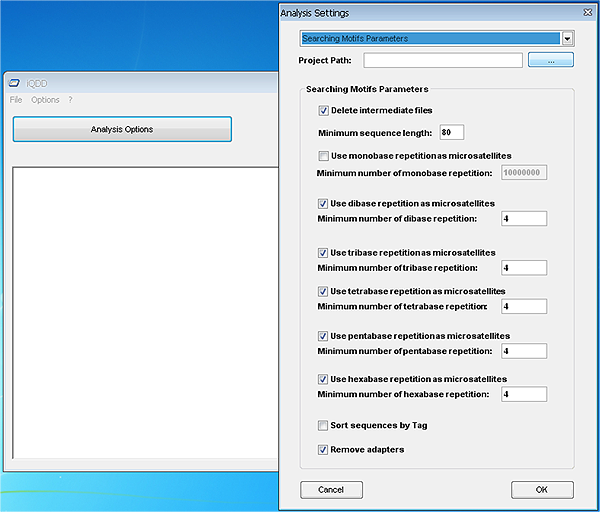 Once the settings are defined, the « Launch Analysis » button is made available and you can run the three steps (searching motifs, checking similarity and redundancy, and primer design). These steps can be run altogether or independently. At the end of the analysis you are proposed to check the results. If you run the whole process, the results (perfect motifs and compound motifs, if selected) are found in the folder yourdataname_pipe3. |
|||
Download QDD1: |
|||
|
Windows
|
Linux
|
||
 QDD_windows (+ iQDD GUI for QDD) (587 ko) QDD_windows (+ iQDD GUI for QDD) (587 ko) |
 QDD_Linux (2.46 Mo) QDD_Linux (2.46 Mo) |
||
 Documentation (2271 ko) Documentation (2271 ko) |
 Documentation (2271 ko) Documentation (2271 ko) |
||
 Sample file for Windows (1167 ko) Sample file for Windows (1167 ko) |
 Sample file for Linux (1209 ko) Sample file for Linux (1209 ko) |
||
Download QDD2: |
|||
| Windows and Linux (Command line only) | |||
 QDD 2.2 Beta QDD 2.2 Beta |
|||
 QDD 2.1 Beta QDD 2.1 Beta |
|||
 Documentation QDD2 Documentation QDD2 |
|||
 Sample files Sample files |
|||
The compression format used for packaging the Windows version was the
RAR file format, most commonly used on DOS/Windows machines.
|
|||
Disclaimer: The software on this page is free to download and use, and thus comes with no warranty of any kind. While it hasn't caused us any problem, the current version of QDD2 is still considered as a beta version and you are responsible for any damages or loss of data you may sustain while using this software. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. Correspondence, comments and bug reports about this program should be addressed to Emese Meglécz |
|||
|
Back to Emese Meglécz's MAIN PAGE Last updated at 16 July 2013
|
|
||